cansas1d casestudy collagen
Overview
To demonstrate how to get SAS data into the XML standard format, consider this set of SAXS data collected at the National Synchrotron Light Source, Brookhaven National Laboratory, using a SAXS camera set up temporarily at beam line X6B (operated by the Materials Science Division, Argonne National Lab).
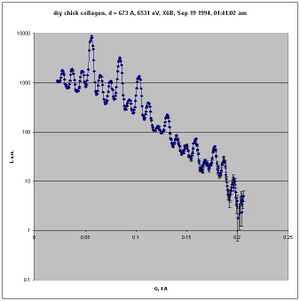
The sample was dry chick collagen.
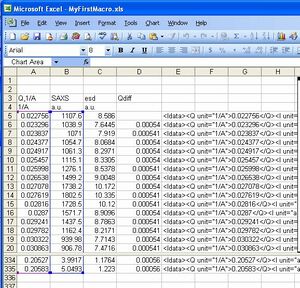
The raw data was collected on a linear position-sensitive detector and reduced to columns of Q, I, and Iesd (estimated standard deviation of I).
The only metadata available for this data (without resorting to digging through piles of old notebooks) was obtained from two file headers:
Sep 19 1994 01:41:02 am Elt: 00090 Seconds ID: No spectrum identifier defined Memory Size: 8192 Chls Conversion Gain: 1024 Adc Offset: 0000 Chls
dry chick collagen, d = 673 A 6531 eV, X6B
But, that will fulfill the minimum requirements of the 1D standard file format and make an excellent example of a minimal canSAS reduced 1-D SAS data file in XML.
Procedure
make the basic XML file
It is easiest to copy a template rather than start from an empty file. Copy the cansas1d.xml file into your working directory and rename it to collagen.xml.
modify collagen.xml
It is easier to see the metadata in the XML file before you enter the SAXS data into the file. With the brief metadata available, most lines can be eliminated. This will result in a file that looks like
<?xml version="1.0"?> <SASroot version="1.0" xmlns="http://www.smallangles.net/cansas1d" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation=" http://www.smallangles.net/cansas1d/ http://www.smallangles.net/cansas1d/1.0/cansas1d.xsd " > <SASentry> <Title>dry chick collagen, d = 673 A, 6531 eV, X6B</Title> <Run /> <SASdata> <!-- Idata lines will go here --> </SASdata> <SASsample> <ID>dry chick collagen, d = 673 A, 6531 eV, X6B</ID> </SASsample> <SASinstrument> <name>X6B, NSLS, BNL</name> <SASsource> <radiation>X-ray synchrotron</radiation> <wavelength unit="A"> 1.898 <!-- = 12398/6531 --> </wavelength> </SASsource> <SAScollimation /> <SASdetector> <name>X6B PSD</name> </SASdetector> </SASinstrument> <SASnote> Sep 19 1994 01:41:02 am Elt: 00090 Seconds ID: No spectrum identifier defined Memory Size: 8192 Chls Conversion Gain: 1024 Adc Offset: 0000 Chls dry chick collagen, d = 673 A 6531 eV, X6B </SASnote> </SASentry> </SASroot>